JMLS 2022 December;7(2):121-128. 10.23005/ksmls.2022.7.2.121 Epub 2022 December 14
Copyright © 2022 by The Korean Society of Marine Life Science
Characterization of L-asparaginase-producing Trichoderma spp. Isolated from Marine Environments
Woon-Jong Yu; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
Dawoon Chung; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
Yong Min Kwon; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
Seung Sub Bae; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
Eun-Seo Cho; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
Hye Suck An; Marine BioIndustrial Research Division, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
Grace Choi; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
- Abstract
L-asparaginase (ASNase) is a therapeutic enzyme used to treat acute lymphoblastic leukemia. Currently, the most widely used ASNases are originated from bacteria. However, owing to the adverse effects of bacterial ASNases, new resources for ASNase production should be explored. Fungal enzymes are considered efficient and compatible resources of natural products for diverse applications. In particular, fungal species belonging to the genus Trichoderma are well-known producers of several commercial enzymes including cellulase, chitinase, and xylanase. However, enzyme production by marine-derived Trichoderma spp. remains to be elucidated. While screening for extracellular ASNase-producing fungi from marine environments, we found four strains showing extracellular ASNase activity. Based on the morphological and phylogenetic analyses using sequences of translation elongation factor 1-alpha (tef1α), the Trichoderma isolates were identified as T. afroharzianum, T. asperellem, T. citrinoviride, and Trichoderma sp. 1. All four strains showed different ASNase activities depending on the carbon sources. T. asperellem MABIK FU00000795 showed the highest ASNase value with lactose as a carbon source. Based on our findings, we propose that marine-derived Trichoderma spp. are potential candidates for novel ASNase production.
Keywords: L-asparaginase Marine fungi Trichoderma spp.
Correspondence to: Grace Choi; Department of Microbial Resources, National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
- Received
- 16 October 2022;
- Revised
- 31 October 2022;
- Accepted
- 22 November 2022.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Language: Korean/English,
Full Text:

Introduction
The genus Trichoderma Persoon belongs to the family Hypoc- reaceae and contains more than 250 species (Bissett et al., 2015). Trichoderma species are ubiquitous and have been isolated from various environments and substrates, such as soil, agricultural fields, and plant debris; as well as in marine resources, including mudflats, sea sand, and seaweeds (Kredics et al., 2014; Kim et al., 2020). Approximately 78 metabolites from Trichoderma spp. have been reported (Su et al., 2018), some of which exhibit antimicrobial, antioxidant, and cytotoxic activities (Zhang et al., 2017; Vinale et al., 2016).
Trichoderma is also known to produce various enzymes, such as an array of carbohydrate-active enzymes (CAZymes). Cellulases, hemicellulases, and chitinases, which belongs to glycoside hydro- lases (GHs), have been employed in the bioconversion of biomass(di Cologna et al., 2018; Peciulyte et al., 2014). For example, T. reesei is a major source of cellulolytic enzymes and is a ligno- cellulosic biomass degrader (Adav et al., 2012; Bischof et al., 2016). T. harzianum has potential biotechnological applications because of its ability to degrade and metabolize lignocellulosic plant materials (Ferreira et al., 2017). In addition, the influence of carbon sources and metals on enzyme production such as cellulase and β-glucosidase in Trichoderma spp. have been studied to under- stand their mechanisms with regard to the induction and regu- lation of enzymes (Mandels and Reese, 1957; Loewenberg, 1984).
L-asparaginase (ASNase), which converts L-asparagine to aspartic acid, is used to treat acute lymphoblastic leukemia (ALL). ALL is the most common cancer in children aged 2~10 years (Egler et al., 2016). In addition, ASNase is used to remove carcinogenic compound acrylamide found in food (Doriya and Kumar, 2016). ASNase is produced by a variety of plants and microorganisms including bacteria, fungi, and algae. These microorganisms, espe- cially bacteria, are relatively inexpensive and efficient sources of ASNase. Indeed, ASNase from Pectobacterium carotovorum and Escherichia coli have been utilized to treat ALL (Ramya et al., 2012). However, due to the side effects induced by bacterial ASNases, such as allergic reactions, fever, and neurological problems, novel ASNases should be explored.
Although the genus Trichoderma is well known for producers of valuable metabolites, production of ASNase by Trichoderma spp. has rarely been reported. Therefore, in this study, we searched for marine-derived Trichoderma spp. with ASNase activity and identified them using morphological and phylogenetic analyses. The effect of carbon sources on the extracellular ASNase activity of Trichoderma spp. was investigated. Through this paper, we suggested Trichoderma spp. served as ASNase sources potentially.
Materials and Methods
1. Isolation and preparation of Trichoderma spp. cultures
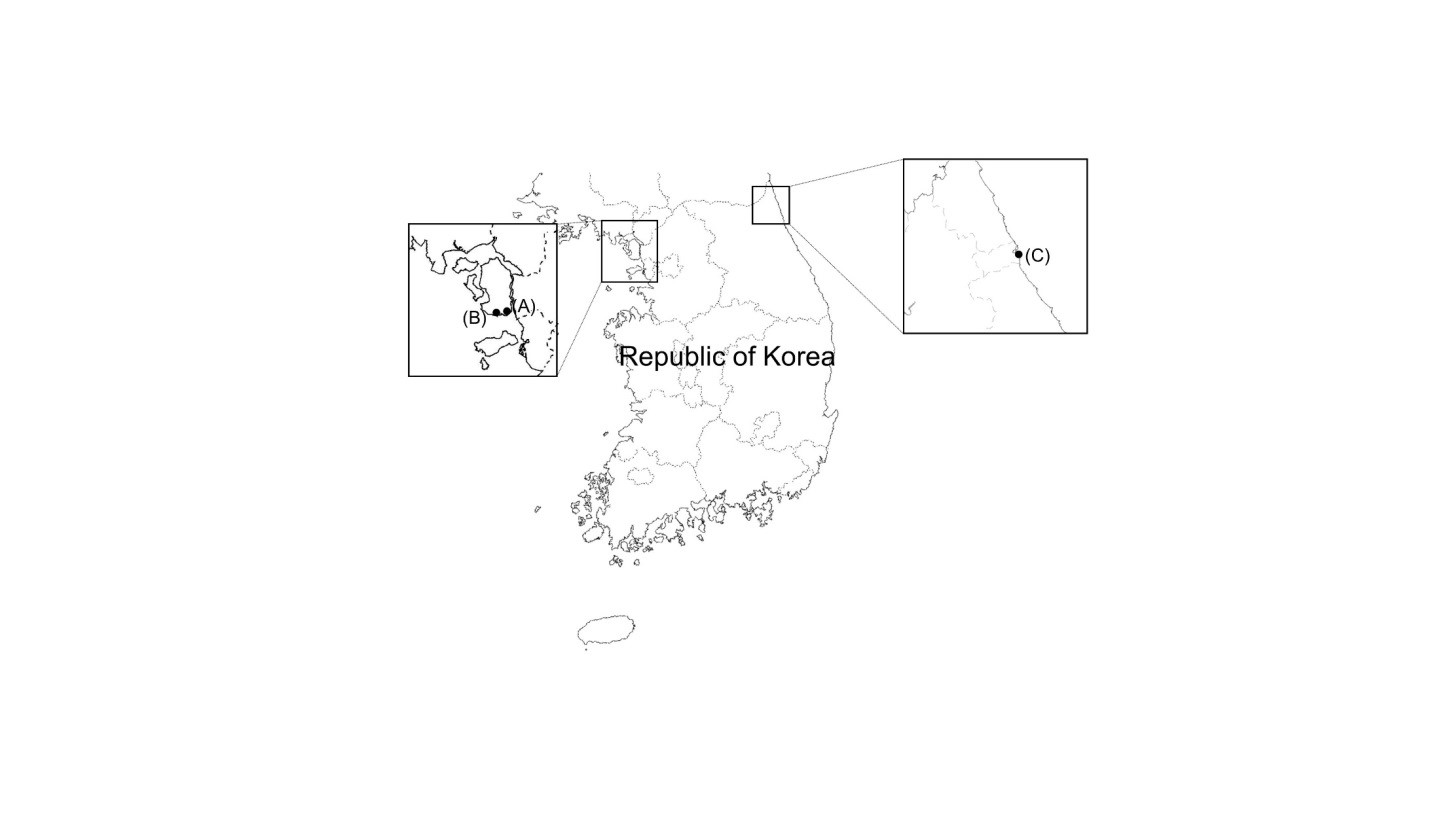
Mudflats and sea sand were collected from Incheon and Sokcho-si, Republic of Korea, and diluted with sterile distilled water. The diluted samples were spread on potato dextrose agar (PDA; BD Difco, Sparks, MD, USA) containing 0.01% (w/v) ampicillin (Sigma-Aldrich, St. Louis, MO, USA) and 0.01% (w/v) streptomycin (Sigma-Aldrich), and incubated at 25℃ for 7~14 days. During that period, individual fungal colonies were picked and transferred to fresh PDA in order to obtain pure cultures. After isolation, fungal strains were cultured on PDA at 28℃ unless otherwise described. Fungal spores and hyphal fractions were suspended in 20% glycerol (v/v) and stored at -80℃. Sample collection and general information for Trichoderma isolates are presented in Table 1 and Fig. 1.
2. Morphological observation
To observe the fungal colony morphology, all isolates were pre-cultured on PDA at 28℃ for 2 days. Subsequently, using agar plugs (8 mm diameter) obtained from the pre-cultures on PDA, individual strains were inoculated on PDA, yeast mold agar (YMA; BD Difco), malt extract agar (MEA; BD Difco), and czapek yeast extract agar (CYA; czapek dox broth; BD Difco, 5.0 g/l yeast extract and 15.0 g/l agar), followed by cultivation at 28℃ for 5 days. Colony characteristics such as color and shape were observed by naked eyes.
3. DNA extraction, sequencing, and phylogenetic analysis
To extract genomic DNA, Trichoderma spp. were incubated in potato dextrose broth (PDB; BD Difco) at 28℃ for 3 days. Genomic DNA was then extracted from the harvested mycelia previously established protocols (Chung et al., 2019). Polymerase chain re- action (PCR) was conducted using the primers EF1-728F (5'-CAT CGA GAA GTT CGA GAA GG-3') (Carbone and Kohn, 1999) and TEF1LLErev (5'-AAC TTG CAG GCA ATG TGG-3') (Jaklitsch et al., 2005) to amplify the translation elongation factor 1-alpha (tef1α) gene. PCR conditions included an initial denaturation of 95℃ for 5 min; followed by 34 cycles of denaturation at 95℃ for 15s, annealing at 58°C for 30s, and extension at 72℃ for 1 min; and finally a post-polymerization at 72℃ for 15 min. PCR products were purified using a Gel Extraction kit (Qiagen, Hilden, Germany). Subsequently, the purified products were sequenced by Macrogen Inc. (Seoul, South Korea).
The tef1α sequences were identified in the GenBank database by BLAST searches. The sequences were aligned and a phylo- genetic tree was constructed using MEGA7.0 (Kumar et al., 2016). A phylogenetic analysis was performed using the neighbor-joining method (Kimura two-parameter model), followed by bootstrapping with 1,000 random replicates (Kimura, 1980) in MEGA7.0. The reference tef1α sequences were obtained from GenBank database.
The four Trichoderma spp. isolates were deposited in Marine Microbial BioBank (MMBB) culture collections of the National Marine Biodiversity Institute of Korea (MABIK) after identification.
4. Screening of L-asparaginase producing Tricho- derma spp.
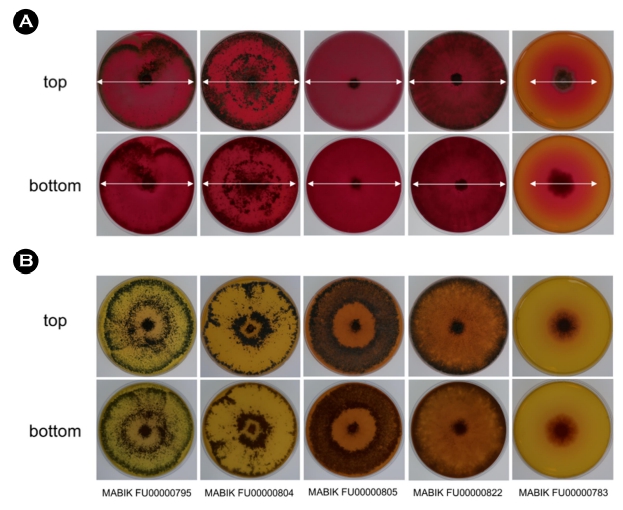
To screen ASNase-producing Trichoderma spp., Modified Czapek Dox agar (MCDA) (Gulati et al., 1997) was used for the plate assay. The components of 1 ℓ of MCDA were as follows: 2.0 g glucose, 10.0 g L-asparagine, 1.52 g KH2PO4, 0.52 g KCl, 0.52 g MgSO4 · 7H2O, 0.05% (w/v) CuNO3 · 3H2O, 0.05% (w/v) ZnSO4 · 7H2O, 0.05% (w/v) FeSO4 · 7H2O and 15.0 g agar, supplemented with 0.005% (w/v) phenol red as indicator of ammonia production. The pH was adjusted to 6.0 using 1 N NaOH. Furthermore, control plates were prepared with NaNO3, instead of L-asparagine, as a nitrogen source. Aspergillus terreus MABIK FU00000783 obtained from MMBB was used as the positive control (de-Angeli et al., 1970). Agar plugs (8 mm diameter) from pre-cultured isolates were inoculated on ASNase-containing and control plates, followed by incubation at 28°C for 5 days. The presence of pink zones around colonies indicated ASNase enzyme activity.
5. Effects of carbon source on extracellular L-asparaginase activities
Agar plugs (diameter 8 mm) of Trichoderma isolates were inoculated in 100 ml of Modified Czapek Dox broth (MCDB) and incubated at 28℃ for 5 days with shaking at 120 rpm. The components of the MCDB were the same as those of the MCDA, except for agar and phenol red. To examine the effect of ASNase production depending on different carbon sources, 0.2% (w/v) glucose, fructose, sucrose, lactose, and soluble starch were supple- mented to MCDB as the sole carbon source. Supernatants from the four isolates were prepared to investigate extracellular ASNase activity. Briefly, the supernatants were collected using a Miracloth (Merck, New Jersey, USA) and sterilized using syringe filters with 0.2 µm pore size (Corning, NY, USA). ASNase activity was meas- ured using the Asparaginase Activity Assay Kit (ab107922, Abcam, Cambridge, MA, USA), according to manufacturer's instructions (Kim et al., 2015). In the assay, fungal supernatants were used to hydrolyze L-asparagine as a substrate to produce L-aspartate, which is subsequently converted to pyruvate. Pyruvate reacts with the probe to form a stable chromophore, which can be detected at 570 nm. One unit of asparaginase was defined as the amount of asparaginase generating 1.0 µmol of aspartate per minute at 25℃. An enclosed positive control solution, which is an asparaginase enzyme solution, was used to ensure confidence in the results.
For data analysis, two-way ANOVA was used to clarify the effects of species and carbon sources on ASNase activities.
Results and Discussion
1. Colony morphology and growth rate of Tricho- derma spp. on various media
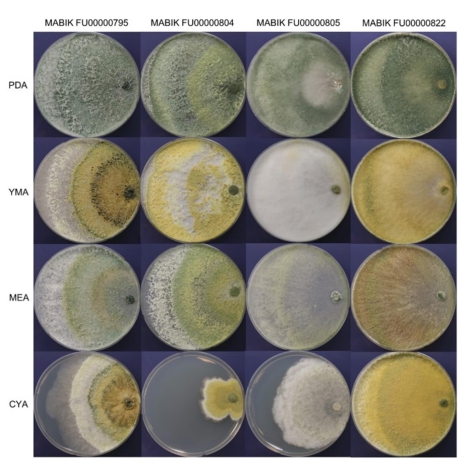
The morphological characteristics of the four Trichoderma strains on PDA, YMA, MEA and CYA were observed (Fig. 2). Strain MABIK FU00000795 formed abundant aerial mycelia and conidia. Its colonies were distributed radically on all types of media. Com- pared to the white to dark green colonies on PDA, the colonies on YMA, MEA and CYA zonated distinctly with different colors. Strain MABIK FU00000804 had abundant conidia, with broad con- centric rings on PDA, YMA and MEA. Its colonies on PDA formed pale olive to pale green conidia, whereas those on YMA formed yellow to grayish yellow pustules. On MEA, colonies zonated dis- tinctly with white and olive-colored conidia in the outermost layer and inner layers, respectively. Furthermore, MABIK FU00000804 had a slow growth on CYA compared on PDA, YMA and MEA, and it had olive-colored conidia around the inoculums. Strain MABIK FU00000805 appeared white and pale green on PDA. Aerial mycelia and cottony conidia were abundant on YMA. On CYA, whitish to pale greenish aerial mycelia appeared hairy to floccose and radical. On all media, strain MABIK FU00000822 formed aerial mycelia that had abundant cottony conidia. Colonies appeared circular, pale green to dark green, and radially distributed on PDA. Meanwhile, yellow-green colonies were observed on YMA, white to olive colonies on MEA, and yellow to dark green colonies on CYA. Diffusing pigments were observed on all media. The pig- ments were yellow to pale green, orange to brown, yellow to pale green, and brown on PDA, YMA, MEA, and CYA, respectively.
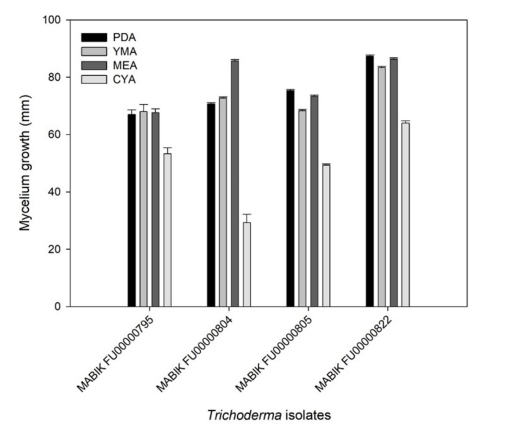
Overall, the four Trichoderma spp. showed distinct growths on PDA, YMA, MEA and CYA (Fig. 3). The growth of mycelia on CYA was slower than that on other media. Taken together, the marine-derived Trichoderma spp. in this study presented different growth rates and colony morphologies depending on the media.
2. Phylogenetic analysis
Morphological and molecular methods have been used to investigate the taxonomy of Trichoderma (Samuels et al., 2002). Distinguishing Trichoderma species based only on morphological characteristics is difficult because they are morphologically similar (Kim et al., 2020). Because the internal transcribed spacer rDNA, the universal fungal DNA barcoding marker, is not sufficient to identify certain fungal clades using molecular methods, tef1α is recommended as a barcoding marker for the phylogeny of Trichoderma (Raja et al., 2017).
In this study, marine-derived Trichoderma spp. were identified via phylogenetic analysis of tef1α (Fig. 4). For molecular identification, the tef1α sequence information were deposited in GenBank (accession numbers were represented in Table 1). The phylogenetic tree contained 36 taxa of Trichoderma strains. The four strains (in bold) were grouped into distinct clades: MABIK FU00000795 in the Hamatum clade, MABIK FU00000804 in the Longibrachiatum clade, and MABIK FU00000805 and MABIK FU- 00000822 in the Harzianum clade. The BLASTN search conducted using tef1α sequences showed that MABIK FU00000795 is closely related to T. asperellum B1902, C261s, T2007, M01, and CBS 433.97. MABIK FU0000084 was grouped with T. citrinoviride DAOM 172792, 18CPHA003, S279 and TRS745. MABIK FU00000795 and MBIK FU00000804 were identified as T. asperellum and T. citrinoviride, respectively, with high bootstrap value of 99% and 100%, respect- ively. MABIK FU00000805 showed a high degree of similarity to T. afroharzianum Z19, Tafum1, and HZA3 (99.48%, 99.65%, and 99.65% identity), but the bootstrap value was relatively low (74%). As MABIK FU00000822 was grouped with two distinct species, T. vermifimicola CGMCC 3.19850 (99.59% identity) and T. xixiacum CGMCC 3.19697 (99.68% identity), it was not identified at the species level and thus was designated as Trichoderma sp. 1.

3. Screening of L-asparaginase-producing Tricho- derma spp.
While we screened extracellular ASNase-producing fungi isolated from marine environments, results of the plate assay indicated four Trichoderma spp. produced ASNase. Trichoderma isolates were cultured on MCDA with L-asparagine as substrate. ASNase activity is accompanied with an increase in the pH due to the pro- duction of aspartic acid and ammonia from L-asparagine. Phenol red as the indicator of alkaline pH represented pink zones on the agar plates. The positive control A. terreus MABIK FU00000783 exhibited ASNase activity, as determined by the presence of a pink zone around the colony on the MCDA containing asparagine. In contrast, inoculation of A. terreus MABIK FU00000783 on the control plate containing NaNO3 as the sole nitrogen source did not produce pink zones. Similarly, all Trichoderma isolates showed pink zones around colonies when grown on asparagine-containing MCDA, whereas no pink zones were observed in the control plates containing NaNO3 (Fig. 5). Therefore, we found ASNase-producing fungal strains that showed a pink zone of the colony. Among these Trichoderma spp., ASNase production using the plate assay method and the characteristics of purified ASNase of T. asperellum have been reported (Uzma et al., 2016; Elsaba et al., 2022). These results indicated that four Trichoderma isolates from marine-associated habitats produced extracellular ASNase and showed ASNase activity.
4. Effects of carbon sources on extracellular L-asparaginase activity
Carbon sources stimulate cellular growth, leading to increased enzyme production (El-Naggar et al., 2019). In this study, we evaluated the effects of carbon sources on extracellular ASNase activity. Trichoderma isolates in MCDB with various carbon sources exhibited different ASNase activities. At 0.5% concentration in a growth medium, glucose could repress ASNase production in certain microorganisms, including Escherichia coli (Garaev and Golub, 1977; Baskar and Renganathan, 2011). Therefore, to avoid this potential adverse effect of carbon sources, we added 0.2% of carbon sources in this study.
In T. asperellum MABIK FU00000795, the highest value of ASNase activity were recorded at 1.51 ± 0.20 U ml-1 with lactose as a carbon source (Fig. 6). The values of ASNase activities of MABIK FU00000795 were recorded at 0.89 ± 0.05 U ml-1 and 1.02 ± 0.11 U ml-1 with glucose and sucrose as the sole carbon source, respectively. In contrast, ASNase activities of T. afroharzianum MABIK FU00000805 and Trichoderma sp. 1 MABIK FU00000822 were the highest with glucose as a carbon source. The ASNase activity of T. citrinoviride MABIK FU00000804 was not significantly different depending on the carbon sources. Additionally, the effects of species and carbon sources on ASNase activities were determined based on two-way ANOVA (p < 0.001).
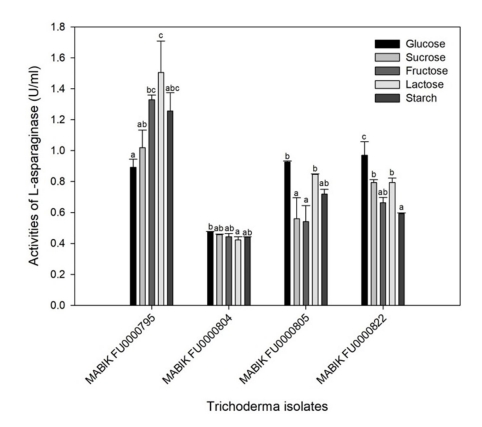
The effects of glucose on ASNase activity vary among the micro- organisms. Glucose can lower the enzyme yield as a catabolic repressor (Heinemann and Howard, 1969), and thus generally serves as a poor carbon source for the production of ASNase in some microorganisms (Lincoln et al., 2021; Freitas et al., 2021; Mukherjee et al., 2000). However, glucose serves as a good carbon source for production of ASNase in some microorganisms such as Aspergillus, Fusarium and Streptomyces strains (Baskar and Renganathan, 2012; Hosamani and Kaliwal, 2011; Deshpande et al., 2014). Therefore, it is important to optimize carbon sources for the production of microbial ASNase.
Conclusion
In this study, we aimed to find novel microbial resources for ASNase production. Four Trichoderma isolates, which were iden- tified using tef1α sequences, were screened for extracellular ASNase activity. T. asperellum MABIK FU0000795 showed the highest ASNase production, with lactose as the sole carbon source. Other carbon sources such as glucose and sucrose served as repressors of the ASNase production of MABIK FU00000795. In contrast, glucose stimulated the enzymatic production of T. afro- harzianum MABIK FU000805 and Trichoderma sp. 1 MABIK FU- 00000822. This study showed ASNase activity of Trichoderma spp. isolated from marine-derived sources. Our results suggest the potential of Trichoderma spp. as ASNase producers. The discovery of novel ASNase producers from eukaryotic microorganisms such as Trichoderma species may be an alternative source with fewer side effects due to their capacity to mimic the characteristics of human cells.
- References
-
1. Adav SS, Chao LT, Sze SK. 2012. Quantitative secretomic analysis of Trichoderma reesei strains reveals enzymatic composition for lignocellulosic biomass degradation. Mol Cell Proteom PM111.012419-1-M111.012419-15.
-
2. Baskar G, Renganathan S. 2011. Optimization of media com- ponents and operating conditions for exogenous production of fungal L-asparaginase. Chiang Mai J Sci 38: 270-279.
-
3. Baskar G, Renganathan S. 2012. Optimization of L‐asparaginase production by Aspergillus terreus MTCC 1782 using response surface methodology and artificial neural network‐linked genetic algorithm. Asia-Pac J Chem Eng 7: 212-220.
-
4. Bischof RH, Ramoni J, Seiboth B. 2016. Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb Cell Factories 15: 1-13.
-
5. Bissett J, Gams W, Jaklitsch W, Samuels GJ. 2015. Accepted Trichoderma names in the year 2015. IMA Fungus 6: 263-295.
-
6. Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553-556.
-
7. Chung D, Baek K, Bae SS, Jung J. 2019. Identification and character- ization of a marine-derived chitinolytic fungus, Acremonium sp. YS2-2. J Microbiol 57: 372-380.
-
8. de-Angeli LC, Pocchiari F, Russi S, Tonolo A, Zurita VE, Ciaranfi E, Perin A. 1970. Effect of L-asparaginase from Aspergillus terreus on ascites sarcoma in the rat. Nature 225: 549-550.
-
9. Deshpande N, Choubey P, Agashe M. 2014. Studies on optimi- zation of growth parameters for L-asparaginase production by Streptomyces ginsengisoli. Sci World J 895167.
-
10. di Cologna NdM, Gómez-Mendoza DP, Zanoelo FF, Giannesi GC, de Alencar Guimarães NC, de Souza Moreira LR, Ferreira Filho EX, Ricart CAO. 2018. Exploring Trichoderma and Aspergillus secretomes: proteomics approaches for the identification of enzymes of biotechnological interest. Enzyme Microb Technol 109: 1-10.
-
11. Doriya K, Kumar DS. 2016. Isolation and screening of L-asparaginase free of glutaminase and urease from fungal sp. 3. Biotech 6: 1-10.
-
12. Egler RA, Ahuja SP, Matloub Y. 2016. L-asparaginase in the treatment of patients with acute lymphoblastic leukemia. J Pharmacol Pharmacother 7: 62.
-
13. El-Naggar NE-A, Moawad H, El-Shweihy NM, El-Ewasy SM, Elsehemy IA, Abdelwahed NA. 2019. Process development for scale-up production of a therapeutic L-asparaginase by Streptomyces brollosae NEAE-115 from shake flasks to bio- reactor. Sci Rep 9: 1-18.
-
14. Elsaba YM, Salama WH, Soliman ER. 2022. In silico and biochemical analysis on a newly isolated Trichoderma asperellum L-asparaginase. Biocatal Agric Biotechnol 2022 40: 102309.
-
15. Ferreira Filho JA, Horta MAC, Beloti LL, Dos Santos CA, de Souza AP. 2017. Carbohydrate-active enzymes in Trichoderma harzi- anum : a bioinformatic analysis bioprospecting for key en- zymes for the biofuels industry. BMC Genom 18: 1-12.
-
16. Freitas M, Souza P, Cardoso S, Cruvinel K, Abrunhosa LS, Ferreira Filho EX, Inácio J, Pinho DB, Pessoa A, O Magalhães P. 2021. Filamentous Fungi Producing L-Asparaginase with Low Glu- taminase Activity Isolated from Brazilian Savanna Soil. Phar- maceutics 13: 1268.
-
17. Garaev M, Golub E. 1977. Mechanism of action of glucose on L-asparaginase synthesis by Escherichia coli bacteria. Mikro- biologiia 46: 433-439.
-
18. Gulati R, Saxena R, Gupta R. 1997. A rapid plate assay for screen- ing L‐asparaginase producing micro‐organisms. Lett Appl Microbiol 24: 23-26.
-
19. Heinemann B, Howard AJ. 1969. Production of tumor-inhibitory L-asparaginase by submerged growth of Serratia marcescens. Appl Microbiol 18: 550-554.
-
20. Hosamani R, Kaliwal B. 2011. L-asparaginase-an anti-tumor agent production by Fusarium equiseti using solid state fermen- tation. Int J Drug Discov 3: 88-99.
-
21. Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. 2005. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97: 1365-1378.
-
22. Kim K, Heo YM, Jang S, Lee H, Kwon SL, Park MS, Lim YW, Kim JJ. 2020. Diversity of Trichoderma spp. in marine environments and their biological potential for sustainable industrial appli- cations. Sustainability 12: 4327.
-
23. Kim K, Jeong JH, Lim D, Hong Y, Lim HJ, Kim GJ, Shin Sr, Lee JJ, Yun M, Harris RA. 2015. L-Asparaginase delivered by Salmonella typhimurium suppresses solid tumors. Mol Ther Oncolytics 2: 15007.
-
24. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111-120.
-
25. Kredics L, Hatvani L, Naeimi S, Körmöczi P, Manczinger L, Vágvölgyi C, Druzhinina I. 2014. Biodiversity of the genus Hypocrea/Trichoderma in different habitats. Gupta V, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy M (ed.), Biotechnology and biology of Trichoderma, Elsevier, Amsterdam, Netherlands, pp 3-24.
-
26. Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolu- tionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874.
-
27. Lincoln L, Niyonzima FN, More SS. 2021. Purification and pro- perties of a fungal L-asparaginase from Trichoderma viride pers: SF GREY. J Microbiol Biotechnol Food Sci 310-316.
-
28. Loewenberg J. 1984. Sophorose induction of an intracellular β-glucosidase in Trichoderma. Arch Microbiol 137: 53-57.
-
29. Mandels M, Reese ET. 1957. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J Bacteriol 73: 269-278.
-
30. Mandels M, Reese ET. 1957. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J Bacteriol 73: 269-278.
-
31. Peciulyte A, Anasontzis GE, Karlström K, Larsson PT, Olsson L. 2014. Morphology and enzyme production of Trichoderma reesei Rut C-30 are affected by the physical and structural char- acteristics of cellulosic substrates. Fungal Genet and Biol 72: 64-72.
-
32. Raja HA, Miller AN, Pearce CJ, Oberlies NH. 2017. Fungal identifi- cation using molecular tools: a primer for the natural products research community. J Nat Prod 80: 756-770.
-
33. Ramya L, Doble M, Rekha V, Pulicherla K. 2012. L-Asparaginase as potent anti-leukemic agent and its significance of having reduced glutaminase side activity for better treatment of acute lymphoblastic leukaemia. Appl Biochem Biotechnol 167: 2144-2159.
-
34. Ramya L, Doble M, Rekha V, Pulicherla K. 2012. L-Asparaginase as potent anti-leukemic agent and its significance of having reduced glutaminase side activity for better treatment of acute lymphoblastic leukaemia. Appl Biochem Biotechnol 167: 2144-2159.
-
35. Su D, Ding L, He S. 2018. Marine-derived Trichoderma species as a promising source of bioactive secondary metabolites. Mini Rev Med Chem 18: 1702-1713.
-
36. Uzma F, Murthy N, Srinivas C. 2016. Optimization of physiological conditions for L-asparaginase production by endophytic fungi (Fusarium solani) isolated from Tinospora cordifolia (Willd.) Hook. F & Thomson. Eur J Exp Biol 6: 37-45.
-
37. Vinale F, Strakowska J, Mazzei P, Piccolo A, Marra R, Lombardi N, Manganiello G, Pascale A, Woo SL, Lorito M. 2016. Cremeno- lide, a new antifungal, 10-member lactone from Trichoderma cremeum with plant growth promotion activity. Nat Prod Res 30: 2575-2581.
-
38. Zhang JC, Chen GY, Li XZ, Hu M, Wang BY, Ruan BH, Zhou H, Zhao LX, Zhou J, Ding ZT. 2017. Phytotoxic, antibacterial, and antioxidant activities of mycotoxins and other metabolites from Trichoderma sp. Nat Prod Res 31: 2745-2752.
-
39. Samuels GJ, Dodd SL, Gams W, Castlebury LA, Petrini O. 2002. Trichoderma species associated with the green mold epi- demic of commercially grown Agaricus bisporus. Mycologia 94: 146-170.
-
40. Mukherjee J, Majumdar S, Scheper T. 2000. Studies on nutritional and oxygen requirements for production of L-asparaginase by Enterobacter aerogenes. Appl Microbiol Biotechnol 53: 180-184.