JMLS 2022 December;7(2):86-93. 10.23005/ksmls.2022.7.2.74 Epub 2022 December 14
Copyright © 2022 by The Korean Society of Marine Life Science
Fluorescent and Luminescent Proteins Derived from Marine Organisms: Functions and Applications
Sehyeok Im; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea, Department of Polar Sciences, University of Science and Technology, Incheon 21990, Korea
Jisub Hwang; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea, Department of Polar Sciences, University of Science and Technology, Incheon 21990, Korea
Hackwon Do; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea, Department of Polar Sciences, University of Science and Technology, Incheon 21990, Korea
Bo-Mi Kim; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea
Sung Gu Lee; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea, Department of Polar Sciences, University of Science and Technology, Incheon 21990, Korea
Jun Hyuck Lee; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea, Department of Polar Sciences, University of Science and Technology, Incheon 21990, Korea
- Abstract
Organisms constituting a large proportion of marine ecosystems, ranging from bacteria to fish, exhibit fluorescence and bioluminescence. A variety of marine organisms utilize these biochemically generated light sources for feeding, reproduction, communication, and defense. Since the discovery of green fluorescent protein and the luciferin-luciferase system more than a century ago, numerous studies have been conducted to characterize their function and regulatory mechanism. The unique properties of fluorescent and bioluminescent proteins offer great potential for their use in a broad range of applications. This short review briefly describes the functions and characteristics of fluorescent and bioluminescent proteins, in addition to summarizing the recent status of their applications.
Keywords: Fluorescence Luminescence Marine Proteins
Correspondence to: Sung Gu Lee; Research Unit of Cryogenic Novel Material, Korea Polar Research Institute, Incheon 21990, Korea
- Received
- 21 September 2022;
- Revised
- 29 September 2022;
- Accepted
- 25 October 2022.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Language: Korean/English,
Full Text:

Introduction
Light produced by living organisms is an attractive subject in multidisciplinary science. Both fluorescence and bioluminescence have been intensively investigated in bacteria, fungi, insects, and marine organisms (Lloyd, 1983; Meyer-Rochow, 2007; Haddock et al., 2010; Widder, 2010). In particular, marine organisms emitting fluorescence or bioluminescence have been intensively investigated for centuries, since the first report on the role of oxygen in bacterial bioluminescence (Boyle, 1667). The habitats of these organisms are broadly distributed on Earth, from the deep sea to the polar regions (Haddock et al., 2010). Luminescence is one of the three major light sources-sunlight, moonlight, and luminescence-for a broad spectrum of marine life, and facilitates a number of their functions, such as those of feeding, reproduction, communication, and defense (Hastings, 1983; Wood et al., 1989; Widder, 1999; Haddock et al., 2009).
Light emissions from living organisms are derived from two distinct systems: fluorescence and bioluminescence. Fluorescence is emitted by a fluorescent photoprotein, wherein a chromophore absorbs light and converts it into a longer wavelength. The green fluorescent protein (GFP) was concomitantly discovered in the jelly- fish Aequorea victoria, when a functional study of the photoprotein aequorin was carried out (Shimomura et al., 1962; Shimomura, 1979). Later investigations identified several members of the GFP superfamily with similar protein structures, although the protein family consists of different protein classes (Yue et al., 2016). Bio- luminescence systems, on the other hand, are more complicated. In general, they require a combination of luciferin and luciferase. The luciferin substrate is activated by the luciferase enzyme, which results in the excited state substrate resuming the ground state and emitting luminescence in the process. To date, only 11 systems of luciferin-luciferase pairs have been elucidated, although more than 30 bioluminescence systems have been identified (Kaskova et al., 2016).
Early studies have revealed the biochemical mechanism of the luciferin-luciferase system and characterized the properties of GFPs (Chiesa et al., 2001; Vysotski and Lee, 2004; Shimomura, 2005). The unique properties of the light-generating systems of these proteins have gathered immense attention in a wide range of multidisciplinary applications, including gene assays, macro- molecule interaction assays, adenosine triphosphate (ATP) deter- mination, biosensors, hygiene control in food industries, in vivo imaging in diagnosis, high-throughput screening in drug develop- ment, and clinical analysis of novel pandemic infectious diseases (Ramsaran et al., 1998; Sylvia et al., 2000; Bhaumik and Gambhir, 2002; Cook and Griffin, 2003; Kadurugamuwa et al., 2003; Cronin et al., 2008; Comps-Agrar et al., 2011; Amodio and Dino, 2014; Lundin, 2014; Karlsson et al., 2015; Lee et al., 2015; Phillips et al., 2016; Taminiau et al., 2016; Belkin et al., 2017; Morciano et al., 2017; Rincon et al., 2017; Iannotti et al., 2018; Dale et al., 2019; Hoare et al., 2019; Endo and Ozawa, 2020; Esteban Florez et al., 2020; Jonkers et al., 2020; Niu et al., 2020; Ong et al., 2020). Marine organisms are enchanting creatures that possess a wide variety of fluorescent and luminescent proteins which have tre- mendous value for a broad range of potential applications (Fig. 1). This review first briefly summarizes the functions of fluorescent and bioluminescent proteins found in marine organisms, then focuses on the recent status of applications of these protein systems, and finally proposes their future perspectives.
Roles of fluorescent proteins in nature
The emission of green fluorescence in the hydrozoan medusa Aequorea victoria depends on the chemiluminescent protein aequorin, which is composed of apoequorin and coelenterazine. Apoaequorin is a 21-kDa single polypeptide that requires Ca2+ and coelenterazine as prosthetic components to release blue light at the wavelength of 470 nm, which is then absorbed by GFP, resulting in the emission of a longer-wavelength green fluor- escent light at the wavelength of 508 nm (Shimomura et al., 1962; Shimomura, 1979). The light conversion is driven by a chromo- phore derived through autocatalytic cyclization of the tripeptide 65-SYG-67 (Shimomura, 1979).
Fluorescent proteins are characterized as having an energy-dispatching function, by means of light scattering, owing to which organisms are capable of protecting themselves against photodamages, such as UVA and radiation (Salih et al., 2000). A previous study demonstrated that reactive oxygen species that occur under the condition of hyperoxia during photosynthesis in algal symbionts are quenched by GFP, thereby suggesting that it has a function of antioxidant protection in A. victoria (Bou-Abdallah et al., 2006). The GFP gene found in cephalochordates has also been proposed to play a role in photoprotection against oxy-radicals (Bomati et al., 2009; Yue et al., 2016). The hydrozoan jellyfish Clytia hemisphaerica possesses four GFPs, and its strong green fluorescence seems to protect stem cells and mitochondrial DNA against UV light (Fourrage et al., 2014).
Fluorescence is attractive to prey, and several studies have identified the function of fluorescent proteins in predation. The fluorescent tentacles of the siphonophore Resomia ornicephala have been shown to serve as prey attractants (Pugh and Haddock, 2010). The GFP in the tentacles of the deep sea anemone Cribri- nopsis japonica absorbs blue light and emits green fluorescence, thereby suggesting its role in prey attraction (Tsutsui et al., 2016). Another group found that the fluorescent proteins in the tentacle tips of the hydromedusa Olindias formosus significantly attracted juvenile rockfish in blue light environments (Haddock and Dunn, 2015).
The intense red fluorescent body pattern observed in the diurnal fish Cirrhilabrus solorensis supports deep-sea vision, and the goby Eviota atriventris is sensitive to red fluorescence (Warrant and Locket, 2004; Michiels et al., 2008). The role of red fluores- cence in vision has been discovered in over 180 species of red fluorescence-emitting marine fish till date (Gerlach et al., 2014; Sparks et al., 2014; Macel et al., 2020). In addition, rays, sharks, and reef fish can display or recognize their own fluorescence, thereby implying that they have the ability to communicate by utilizing it (Heinermann, 1984; Gruber et al., 2016). Research on several copepods and bony fish has suggested other possible roles for fluorescence in mating and camouflage (Shagin et al., 2004; Hunt et al., 2010; Gruber et al., 2016). However, the role of fluorescence in visual recognition among marine organisms remains to be elucidated through further intensive research.
Characteristics of bioluminescent proteins
More than 500 genera dwelling in the ocean, ranging from bacteria to fish, are luminous organisms (Widder, 1999; Haddock et al., 2010; Martini and Haddock, 2017; Shimomura, 2012). Diverse organisms have the ability to visually detect bioluminescence in marine environments, where sunlight or moonlight is poor or unavailable (Widder, 1999, 2002). Daylight declines by nearly 10-fold at every 5 m of depth and completely vanishes below 1,000 m (Widder, 2002). Bioluminescence is considered a major light source in areas where light is rarely available or those that are completely dark. In this context, bioluminescent light is critical for the survival of most deep-sea species, for their hunting, mating, and defense (Widder, 1999; Inouye et al., 2000; Meyer-Rochow, 2007; Stanger-Hall et al., 2007; Haddock et al., 2010).
Since Boyle reported the importance of oxygen in bacterial bioluminescence (Boyle, 1667), other researchers have also investi- gated the mechanisms of light production (Meyer-Rochow, 2007). In this process, Dubois and Harvey noticed the bioluminescence of the click beetle Pyrophorus sp. and marine bivalve mollusc Pholas dactylus and discovered the luciferin-luciferase system (Dubois, 1885; Harvey, 1957; Poisson, 2010; Shimomura, 2012). This finding was then extended to Shimomura's discovery of aequorin, the first bioluminescent photoprotein, in the jellyfish A. victoria (Shimomura et al., 1962). Functional photoproteins are composed of apoproteins, chromophoric components, and oxygen molecules (Ohmiya and Hirano, 1996). The structure of photoproteins is similar among various species that emit bioluminescence, particu- larly in Ca2+-binding sites and spatial structures (Stepanyuk et al., 2013). Interestingly, in several dinoflagellates, the light-emitting activity of luciferase is regulated by a luciferin-binding protein, by sequestering the luciferin substrate at higher pH (Liu et al., 2004).
In a typical luciferin-luciferase system, luciferin is enzymatically oxidized and converted into an excited-state anionic species by luciferase. The excited state oxyluciferin then releases fluorescent blue light at wavelengths in the range of 454~493 nm, following which it returns to its ground state (Henry and Michelson, 1978; Shimomura, 2012). As a typical luciferin, the imidazopyrazine com- pound coelenterazine is oxidized to the excited state coelentera- mide oxyluciferin through a dioxetanone intermediate, resulting in the release of bioluminescence (Shimomura and Johnson, 1978). Unlike insect d-luciferin, coelenterazine is not ATP-dependent for activation (Ohmiya and Hirano, 1996). It was first identified in deep-sea shrimp and copepods (Shimomura et al., 1978; Markova et al., 2019), and is utilized as a substrate for luminescence generation by approximately 15 different luciferases, including the most common luciferases, the sea pansy Renilla reniformis luciferase (Rluc), marine copepod Gaussia princeps luciferase (Gluc), and another marine copepod Metridia longa luciferase (Mluc) (Lorenz et al., 1991; Inouye et al., 2000; Verhaegent and Christopoulos, 2002; Markova et al., 2004; Stepanyuk et al., 2008; Takenaka et al., 2008; Titushin et al., 2008; Takenaka et al., 2012). These luciferases are smaller (~34 kDa Rluc and ~20 kDa Gluc and Mluc) than the luciferase found in terrestrial insects, Fluc (~62 kDa), which makes them appropriate for various applications (Syed and Anderson, 2021).
It is common to observe a similar protein structure for lucif- erases among marine species. For instance, bioluminescent marine dinoflagellates exhibit highly conserved central domains in their luciferases (Liu et al., 2004). Interestingly, the luciferin in the marine ostracod Vargula hilgendorfii showed a cross-reaction with lumi- nescent fish luciferases, thereby giving rise to questions about its evolutionary origin (Thompson et al., 1989). Similar cross-reactivity has also been observed between Euphausiid krill and dinofla- gellates (Nakamura et al., 1988). The discovery of more unknown luciferin-luciferase systems is an active field of research, which will broaden our knowledge of their ecological importance in marine environments and future applications.
Collaborations between fluorescent and bioluminescent proteins
Fluorescence and bioluminescence can exist simultaneously and also interact with the same organism or similar habitats. In this case, bioluminescence acts as a source of light energy for fluorescence generation. For example, when aequorin releases blue luminescent light, this energy is then absorbed by GFP, resulting in the emission of a longer-wavelength green fluorescent light (Shimomura et al., 1962; Shimomura, 1979). Aequorin, therefore, is regarded as a blue fluorescent protein. In Renilla, the blue light released from luciferase Rluc is transferred to the fluorophore of a nearby GFP, resulting in the emission of green fluorescent light at the wavelength of 510 nm (Wang et al., 1998). The siphonophore Erenna sirena converts luminescence generated from its lumi- nescent photophore to fluorescence using tentacles and releases yellow to red light in the wavelength range of 583~680 nm (Haddock et al., 2005). Cnidarians are capable of collecting light energy by fluorescence from the blue luminescence produced in deep-sea habitats (Matz et al., 2006). Interestingly, in several dinoflagellates, luminescent organs are bifunctional. For example, luminescent photophores in the dinoflagellate alga Gonyaulax are considered autofluorescent organs that convert blue luminescence into green light using GFP (Johnson et al., 1985). Considering the light-limited world of the deep sea, it is not surprising to observe interactions between fluorescence and bioluminescence. Conse- quently, it would not be unreasonable to anticipate that there could be further undiscovered events, because the lives in the marine ecosystem are connected to each other for multiple purposes.
Applications
In basic research, fluorescent and bioluminescent proteins are utilized as reporters to trace the expression of specific genes of interest. The recombinant GFP-tagging strategy has been widely adopted for targeting proteins and is now a common tool in molecular biology, cell biology, and biomedicine (Shimomura et al., 1962; Mocz, 2007; Scott et al., 2011). Bioluminescence properties have been widely accepted in the field of applications (Table 1). Early studies adopted bioluminescence resonance energy transfer (BRET) assays for protein-protein interaction studies (Pfleger and Eidne, 2006; England et al., 2016; Dale et al., 2019). In this system, fluorescent and bioluminescent proteins function in a co-operative sequential reaction. An excited-state aequorin or coelenterazine serves as a donor of non-radiative energy and transfers the energy to a proximate acceptor, GFP, followed by the emission of fluorescent light. The proximal distance between the energy donor and acceptor molecules determines the fraction of the energy transfer. This system has been used to study the target proteins of G protein-linked receptors and p53 involved in the oncological progress (Dudgeon et al., 2010; Comps-Agrar et al., 2011). More- over, the recently developed NanoBRET system has been used to identify in vivo protein-ligand interactions using red light-emitting acceptors (Hoare et al., 2019). The split luciferase assay is another major technology that has been used to study protein-protein interactions (Wehr and Rossner, 2016). This system is based on diverse luciferases that are divided into two functional fragments, N-terminal and C-terminal domains, each of which is designed to build a fusion protein of interest. The presence of a ligand recruits the two separate proteins into close proximity, subsequently leading to the reassembly of the two luciferase domains and restoration of the functional protein. A previous study applied Gluc to this assay system, and conducted pharmaceutical validation by visualizing the interaction dynamics (Remy and Michnick, 2006). Luciferase assays have also been used to detect various coronavirus infections in diverse cells (Zhao et al., 2013; Yang et al., 2014). In a study on SARS-Cov-2, genes of both Fluc and the viral spike glycoprotein were introduced into host cells, and the resulting recombinant viral particles containing the bioluminescent reporter Fluc were then used to identify the cellular receptor angiotensin-converting enzyme 2 (Lan et al., 2020; Shang et al., 2020). In other studies, aequorin has been used as a labeling molecule for tumor necrosis factor-alpha, Forssman antigen, cytokine, and protein A (Erikaku et al., 1991; Stults et al., 1992; Xiao et al., 1996; Zatta, 1996). It has also been shown that a limited amount of prostate-specific antigen can be detected by means of an expression immunoassay using aequorin (White and Christopoulos, 1999; Dragulescu-Andrasi et al., 2009; Byun et al., 2019). Several other studies have reported the function of bioluminescence as sensors, for detection of acidosis, reactive oxygen species, nitric oxide, and Ca2+ signaling (Takakura et al., 2015; Pelentir et al., 2019; Ong et al., 2020).
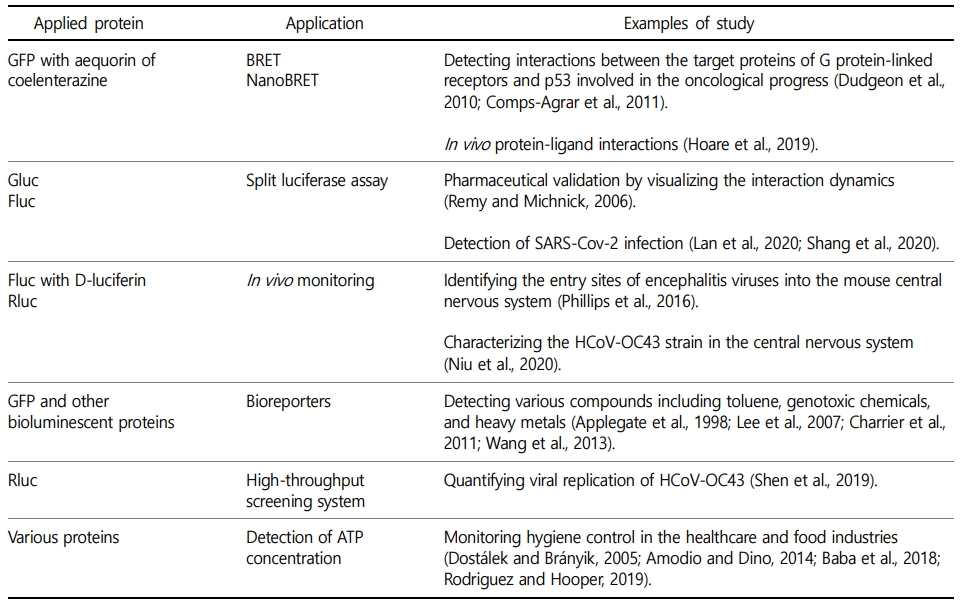
Bioluminescence offers versatile applications for imaging tumor progression (Choy et al., 2003), and has been used in several cancer studies conducted on the breast, colon, and prostate using animal models (Caceres et al., 2003; Scatena et al., 2004; Zeamari et al., 2004). In vivo monitoring is a prominent advantage that bioluminescence offers in laboratory experiments. Light from living cells circumvents the problem of biopsy, which requires that the experimental animals be sacrificed in order to observe the effect of a certain gene expression (Syed and Anderson, 2021). For in- stance, Fluc and D-luciferine have been used to identify the entry sites of encephalitis viruses into the mouse central nervous system (Phillips et al., 2016). In another study, Rluc and coelenterazine were used in live mice, to characterize the HCoV-OC43 strain in the central nervous system (Niu et al., 2020). In the industrial sector, bioluminescence has been utilized in a broad spectrum of applications, with distinct advantages (Syed and Anderson, 2021). The effects of antibiotics have been evaluated over time using animal models. A study genetically engineered target bacteria with the lux operon prior to injection, thus allowing for monitoring of the effect of an antibiotic in vivo in the injected mice (Berger et al., 2017). This advantage can also be applied in photodynamic therapy. In a previous report, bioluminescent bacteria were loaded on the dermal abrasion site of mice treated with photosensitizers, following which the photodynamic therapy efficacy was visually evaluated under red light (Vecchio et al., 2013). There is another interesting possibility for the application of bioluminescent protein as bioreporters. Bioluminescent bioreporters have been shown to effectively detect various compounds including toluene, genotoxic chemicals, and heavy metals (Applegate et al., 1998; Lee et al., 2007; Charrier et al., 2011; Wang et al., 2013). In another study, a recombinant bacterial strain functioning as a trinitrotoluene sensor was constructed by placing a trinitrotoluene-inducible promoter in front of the GFP gene. Sensing for this system was performed using a laser-based optoelectronic system and scanner (Belkin et al., 2017). In the same context, a similar construction of a recombinant strain could be developed for the detection of terrorist com- pounds.
Bioluminescence could be a useful tool in the pharma- ceutical industry, because novel drug discovery demands high-throughput screening systems. In a recent report, Rluc was in- serted into the ns2 accessory gene of the coronavirus strain HCoV-OC43, following which the recombinant strain was trans- fected into BHK-21 cells. The luminescent light emitted from this system upon addition of coelenterazine was then measured to quantify viral replication. Interestingly, this measurement was significantly reduced by a new antiviral drug being tested, thus demonstrating its efficacy. The group successfully tested 2,000-compound libraries using this robust screening system (Shen et al., 2019). In addition, the bioluminescence of insects is suitable for discriminating not only live and dead cells, but also healthy and diseased cells, by utilizing their sensitive detection mechanism of ATP concentration (Zhang et al., 2010; Bird et al., 2014; Lee et al., 2015; Palikaras and Tavernarakis, 2016). This unique feature of the ATP bioluminescence system has been applied to monitor hygiene control in the healthcare and food industries (Dostálek and Brányik, 2005; Amodio and Dino, 2014; Baba et al., 2018; Rodriguez and Hooper, 2019). The respiratory chain of biolumi- nescent bacteria is directly connected to bioluminescence output, and thus, their luminescence system is utilized as a monitoring sensor in ecotoxicology (Fukuba et al., 2011; Hassan et al., 2016; Hansen et al., 2019).
Conclusions and Future perspectives
Even though this review discusses many examples of the applications of marine fluorescence and bioluminescence, a wide spectrum of developments using these stunning lighting machineries are still possible in the future, with active basic studies on these proteins being published regularly. Fluorescent and bio- luminescent proteins possess fascinating properties, which can be functionally enhanced using genetic engineering. For example, mutant luciferases of Rluc have been developed to achieve brighter and more stable enzymes, which have demonstrated enhanced BRET efficiency in in vitro/in vivo imaging and Ca2+ ion-detection ability (Loening et al., 2006; Takai et al., 2015; Suzuki et al., 2016). Moreover, the luciferase from the deep sea caridean shrimp Oplophorus gracilirostris (Oluc) has been engineered by means of mutagenesis to reveal improved light emission and thermal stability (Hall et al., 2012). With the continuous development of functionally enhanced enzymes and substrates, the application area of these proteins could be expanded. Concurrently, the discovery of new luciferin-luciferase and photoprotein systems could result in further enhanced output, by building various combinations of the substrate and enzyme, or fluorescence and luminescence. In addition, Antarctic marine environments have diverse ecosystems, which preserve and offer tremendous potential resources for the development of future biotechnology. Consequently, it is not difficult to anticipate the discovery of novel fluorescent and bio- luminescent proteins in Antarctic marine species from either shallow or deep water, which would contribute to a comprehensive understanding of the entire marine ecosystem as well as the development of beneficial applications for humans.
- References
-
1. Amodio E, Dino C. 2014. Use of ATP bioluminescence for assessing the cleanliness of hospital surfaces: a review of the published literature (1990-2012). J Infect Public Health 7: 92-98.
-
2. Applegate BM, Kehrmeyer SR, Sayler GS. 1998. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, tolu- ene, ethybenzene, and xylene (BTEX) sensing. Appl Environ Microbiol 64: 2730-2735.
-
3. Baba Y, Sato Y, Owada G, Minakuchi S. 2018. Effectiveness of a combination denture-cleaning method versus a mechanical method: comparison of denture cleanliness, patient satis- faction, and oral health-related quality of life. J Prosthodont Res 62: 353-358.
-
4. Belkin S, Yagur-Kroll S, Kabessa Y, Korouma V, Septon T, Anati Y, et al. 2017. Remote detection of buried landmines using a bacterial sensor. Nat Biotechnol 35: 308-310.
-
5. Berger CN, Crepin VF, Roumeliotis TI, Wright JC, Carson D, Pevsner-Fischer M, et al. 2017. Citrobacter rodentium Subverts ATP Flux and Cholesterol Homeostasis in Intestinal Epithelial Cells In vivo. Cell Metab 26: 738-752 e736.
-
6. Bhaumik S, Gambhir SS. 2002. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A 99: 377-382.
-
7. Bird MJ, Wijeyeratne XW, Komen JC, Laskowski A, Ryan MT, Thorburn DR, Frazier AE. 2014. Neuronal and astrocyte dys- function diverges from embryonic fibroblasts in the Ndufs4fky /fky mouse. Biosci Rep 34: e00151.
-
8. Bomati EK, Manning G, Deheyn DD. 2009. Amphioxus encodes the largest known family of green fluorescent proteins, which have diversified into distinct functional classes. BMC Evol Biol 9: 77.
-
9. Bou-Abdallah F, Chasteen ND, Lesser MP. 2006. Quenching of superoxide radicals by green fluorescent protein. Biochim Biophys Acta 1760: 1690-1695.
-
10. Boyle R. 1667. New Experiments concerning the relation between light and air (in shining wood and fish). Philosophical Trans- actions Royal Society 2: 605-612.
-
11. Byun JY, Lee KH, Shin YB, Kim DM. 2019. Cascading Amplification of Immunoassay Signal by Cell-Free Expression of Firefly Luciferase from Detection Antibody-Conjugated DNA in an Escherichia coli Extract. ACS Sens 4: 93-99.
-
12. Caceres G, Zankina R, Zhu X, Jiao JA, Wong H, Aller A, Andreotti P. 2003. Determination of chemotherapeutic activity in vivo by luminescent imaging of luciferase-transfected human tumors. Anticancer Drugs 14: 569-574.
-
13. Charrier T, Durand MJ, Jouanneau S, Dion M, Pernetti M, Poncelet D, Thouand G. 2011. A multi-channel bioluminescent bacterial biosensor for the on-line detection of metals and toxicity. Part I: design and optimization of bioluminescent bacterial strains. Anal Bioanal Chem 400: 1051-1060.
-
14. Chiesa A, Rapizzi E, Tosello V, Pinton P, de Virgilio M, Fogarty KE, Rizzuto R. 2001. Recombinant aequorin and green fluor- escent protein as valuable tools in the study of cell signalling. Biochem J 355: 1-12.
-
15. Choy G, Choyke P, Libutti SK. 2003. Current advances in molecular imaging: noninvasive in vivo bioluminescent and fluorescent optical imaging in cancer research. Mol Imaging 2: 303-312.
-
16. Comps-Agrar L, Maurel D, Rondard P, Pin JP, Trinquet E, Prezeau L. 2011. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to G protein-coupled receptor oligomerization. Methods Mol Biol 756: 201-214.
-
17. Cook SH, Griffin DE. 2003. Luciferase imaging of a neurotropic viral infection in intact animals. J Virol 77: 5333-5338.
-
18. Cronin M, Sleator RD, Hill C, Fitzgerald GF, van Sinderen D. 2008. Development of a luciferase-based reporter system to moni- tor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol 8: 161.
-
19. Dale NC, Johnstone EKM, White CW, Pfleger KDG. 2019. NanoBRET: The Bright Future of Proximity-Based Assays. Front Bioeng Biotechnol 7: 56.
-
20. Dostálek P, Brányik T. 2005. Prospects for rapid bioluminescent detection methods in the food industry – a review. Czech J Food Sci 23: 85-92.
-
21. Dragulescu-Andrasi A, Liang G, Rao J. 2009. In vivo biolumines- cence imaging of furin activity in breast cancer cells using bioluminogenic substrates. Bioconjug Chem 20: 1660-1666.
-
-
23. Dudgeon DD, Shinde SN, Shun TY, Lazo JS, Strock CJ, Giuliano KA, et al. 2010. Characterization and optimization of a novel protein-protein interaction biosensor high-content screening assay to identify disruptors of the interactions between p53 and hDM2. Assay Drug Dev Technol 8: 437-458.
-
24. Endo M, Ozawa T. 2020. Advanced Bioluminescence System for In vivo Imaging with Brighter and Red-Shifted Light Emission. Int J Mol Sci 21.
-
25. England CG, Ehlerding EB, Cai W. 2016. NanoLuc: A Small Lucif- erase Is Brightening Up the Field of Bioluminescence. Bio- conjug Chem 27: 1175-1187.
-
26. Erikaku T, Zenno S, Inouye S. 1991. Bioluminescent immunoassay using a monomeric Fab'-photoprotein aequorin conjugate. Biochem Biophys Res Commun 174: 1331-1336.
-
27. Esteban Florez FL, Hiers RD, Zhao Y, Merritt J, Rondinone AJ, Khajotia SS. 2020. Optimization of a real-time high-throughput assay for assessment of Streptococcus mutans metabolism and screening of antibacterial dental adhesives. Dent Mater 36: 353-365.
-
28. Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791.
-
29. Fourrage C, Swann K, Gonzalez Garcia JR, Campbell AK, Houliston E. 2014. An endogenous green fluorescent protein-photoprotein pair in Clytia hemisphaerica eggs shows co-targeting to mitochondria and efficient bioluminescence energy transfer. Open Biol 4: 130206.
-
30. Fukuba T, Aoki Y, Fukuzawa N, Yamamoto T, Kyo M, Fujii T. 2011. A microfluidic in situ analyzer for ATP quantification in ocean environments. Lab Chip 11: 3508-3515.
-
31. Gerlach T, Sprenger D, Michiels NK. 2014. Fairy wrasses perceive and respond to their deep red fluorescent coloration. Proc Biol Sci 281.
-
32. Gruber DF, Loew ER, Deheyn DD, Akkaynak D, Gaffney JP, Smith WL, et al. 2016. Biofluorescence in Catsharks (Scyliorhinidae): Fundamental Description and Relevance for Elasmobranch Visual Ecology. Sci Rep 6: 24751.
-
33. Haddock SH, Dunn CW. 2015. Fluorescent proteins function as a prey attractant: experimental evidence from the hydromedusa Olindias formosus and other marine organisms. Biol Open 4: 1094-1104.
-
-
-
36. Haddock SH, Dunn CW, Pugh PR, Schnitzler CE. 2005. Biolumi- nescent and red-fluorescent lures in a deep-sea siphono- phore. Science 309: 263.
-
37. Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, et al. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7: 1848-1857.
-
38. Hansen CB, Kerrouche A, Tatari K, Rasmussen A, Ryan T, Summersgill P, et al. 2019. Monitoring of drinking water quality using automated ATP quantification. J Microbiol Methods 165: 105713.
-
39. Harvey EN. 1957. A history of luminescence from the earliest times until 1900. American Philosophical Society. Philadelphia.
-
40. Hassan SH, Van Ginkel SW, Hussein MA, Abskharon R, Oh SE. 2016. Toxicity assessment using different bioassays and microbial biosensors. Environ Int 92-93: 106-118.
-
41. Hastings JW. 1983. Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. J Mol Evol 19: 309-321.
-
-
43. Henry JP, Michelson AM. 1978. Bioluminescence: physiological control and regulation at the molecular level. Photochem Photobiol 28: 293-310.
-
44. Hoare BL, Bruell S, Sethi A, Gooley PR, Lew MJ, Hossain MA, et al. 2019. Multi-Component Mechanism of H2 Relaxin Binding to RXFP1 through NanoBRET Kinetic Analysis. iScience 11: 93-113.
-
45. Hunt ME, Scherrer MP, Ferrari FD, Matz MV. 2010. Very bright green fluorescent proteins from the Pontellid copepod Pontella mimocerami. PLoS One 5: e11517.
-
46. Iannotti FA, Pagano E, Guardiola O, Adinolfi S, Saccone V, Consalvi S, et al. 2018. Genetic and pharmacological regulation of the endocannabinoid CB1 receptor in Duchenne muscular dystrophy. Nat Commun 9: 3950.
-
47. Inouye S, Watanabe K, Nakamura H, Shimomura O. 2000. Secre- tional luciferase of the luminous shrimp Oplophorus gracilir- ostris: cDNA cloning of a novel imidazopyrazinone luciferase. FEBS Lett 481: 19-25.
-
48. Johnson CH, Inoue S, Flint A, Hastings JW. 1985. Compart- mentalization of algal bioluminescence: autofluorescence of bioluminescent particles in the dinoflagellate Gonyaulax as studied with image-intensified video microscopy and flow cytometry. J Cell Biol 100: 1435-1446.
-
49. Jonkers TJH, Steenhuis M, Schalkwijk L, Luirink J, Bald D, Houtman CJ, et al. 2020. Development of a high-throughput bioassay for screening of antibiotics in aquatic environmental samples. Sci Total Environ 729: 139028.
-
50. Kadurugamuwa JL, Sin LV, Yu J, Francis KP, Kimura R, Purchio T, Contag PR. 2003. Rapid direct method for monitoring anti- biotics in a mouse model of bacterial biofilm infection. Anti- microb Agents Chemother 47: 3130-3137.
-
51. Karlsson EA, Meliopoulos VA, Savage C, Livingston B, Mehle A, Schultz-Cherry S. 2015. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat Commun 6: 6378.
-
52. Kaskova ZM, Tsarkova AS, Yampolsky IV. 2016. 1001 lights: lucif- erins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem Soc Rev 45: 6048-6077.
-
53. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35:1547-1549.
-
54. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581: 215-220.
-
55. Lee JH, Youn CH, Kim BC, Gu MB. 2007. An oxidative stress-specific bacterial cell array chip for toxicity analysis. Biosens Bioelectron 22: 2223-2229.
-
56. Lee S, Zhang C, Liu X. 2015. Role of glucose metabolism and ATP in maintaining PINK1 levels during Parkin-mediated mito- chondrial damage responses. J Biol Chem 290: 904-917.
-
57. Liu L, Wilson T, Hastings JW. 2004. Molecular evolution of dinofla- gellate luciferases, enzymes with three catalytic domains in a single polypeptide. Proc Natl Acad Sci U S A 101: 16555-16560.
-
-
59. Loening AM, Fenn TD, Wu AM, Gambhir SS. 2006. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel 19: 391-400.
-
60. Lorenz WW, McCann RO, Longiaru M, Cormier MJ. 1991. Isolation and expression of a cDNA encoding Renilla reniformis lucif- erase. Proc Natl Acad Sci U S A 88: 4438-4442.
-
61. Lundin A. 2014. Optimization of the firefly luciferase reaction for analytical purposes. Adv Biochem Eng Biotechnol 145: 31-62.
-
62. Macel ML, Ristoratore F, Locascio A, Spagnuolo A, Sordino P, D'Aniello S. 2020. Sea as a color palette: the ecology and evolution of fluorescence. Zoological Lett 6: 9.
-
63. Markova SV, Larionova MD, Vysotski ES. 2019. Shining Light on the Secreted Luciferases of Marine Copepods: Current Know- ledge and Applications. Photochem Photobiol 95: 705-721.
-
64. Markova SV, Golz S, Frank LA, Kalthof B, Vysotski ES. 2004. Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa. A novel secreted biolumi- nescent reporter enzyme. J Biol Chem 279: 3212-3217.
-
65. Martini S, Haddock SH. 2017. Quantification of bioluminescence from the surface to the deep sea demonstrates its predom- inance as an ecological trait. Sci Rep 7: 45750.
-
66. Matz MV, Labas YA, Ugalde J. 2006. Evolution of function and color in GFP-like proteins. Methods Biochem Anal 47: 139-161.
-
67. Meyer-Rochow VB. 2007. Glowworms: a review of Arachnocampa spp. and kin. Luminescence 22: 251-265.
-
68. Michiels NK, Anthes N, Hart NS, Herler J, Meixner AJ, Schleifenbaum F, et al. 2008. Red fluorescence in reef fish: a novel signalling mechanism? BMC Ecol 8: 16.
-
69. Mocz G. 2007. Fluorescent proteins and their use in marine bio- sciences, biotechnology, and proteomics. Mar Biotechnol (NY) 9: 305-328.
-
70. Morciano G, Sarti AC, Marchi S, Missiroli S, Falzoni S, Raffaghello L, et al. 2017. Use of luciferase probes to measure ATP in living cells and animals. Nat Protoc 12: 1542-1562.
-
71. Nakamura H, Musicki B, Kishi Y, Shimomura O. 1988. Structure of the light emitter in krill (Euphausia pacifica) bioluminescence. J Am Chem Soc 110: 2683-2685.
-
72. Niu J, Shen L, Huang B, Ye F, Zhao L, Wang H, et al. 2020. Non-invasive bioluminescence imaging of HCoV-OC43 infection and therapy in the central nervous system of live mice. Antiviral Res 173: 104646.
-
73. Ohmiya Y, Hirano T. 1996. Shining the light: the mechanism of the bioluminescence reaction of calcium-binding photoproteins. Chem Biol 3: 337-347.
-
74. Ong TT, Ang Z, Verma R, Koean R, Tam JKC, Ding JL. 2020. pHLuc, a Ratiometric Luminescent Reporter for in vivo Monitoring of Tumor Acidosis. Front Bioeng Biotechnol 8: 412.
-
75. Palikaras K, Tavernarakis N. 2016. Intracellular Assessment of ATP Levels in Caenorhabditis elegans. Bio Protoc 6.
-
76. Pelentir GF, Bevilaqua VR, Viviani VR. 2019. A highly efficient, thermostable and cadmium selective firefly luciferase suitable for ratiometric metal and pH biosensing and for sensitive ATP assays. Photochem Photobiol Sci 18: 2061-2070.
-
77. Pfleger KD, Eidne KA. 2006. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat Methods 3: 165-174.
-
78. Phillips AT, Rico AB, Stauft CB, Hammond SL, Aboellail TA, Tjalkens RB, Olson KE. 2016. Entry Sites of Venezuelan and Western Equine Encephalitis Viruses in the Mouse Central Nervous System following Peripheral Infection. J Virol 90: 5785-5796.
-
79. Poisson J. 2010. [Raphael Dubois, from pharmacy to biolumi- nescence]. Rev Hist Pharm (Paris) 58: 51-56.
-
80. Pugh PR, Haddock SH. 2010. Three new species of remosiid siphonophore (Siphonophora: Physonectae). J Mar Biol Assoc United Kingdom 90: 1119-1143.
-
81. Ramsaran H, Chen J, Brunke B, Hill A, Griffiths MW. 1998. Survival of bioluminescent Listeria monocytogenes and Escherichia coli O157:H7 in soft cheeses. J Dairy Sci 81: 1810-1817.
-
82. Remy I, Michnick SW. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods 3: 977-979.
-
83. Rincon E, Cejalvo T, Kanojia D, Alfranca A, Rodriguez-Milla MA, Gil Hoyos RA, et al. 2017. Mesenchymal stem cell carriers enhance antitumor efficacy of oncolytic adenoviruses in an immunocompetent mouse model. Oncotarget 8: 45415-45431.
-
84. Rodriguez JA, Hooper G. 2019. Adenosine Triphosphate-Biolumi- nescence Technology as an Adjunct Tool to Validate Clean- liness of Surgical Instruments. AORN J 110: 596-604.
-
85. Salih A, Larkum A, Cox G, Kuhl M, Hoegh-Guldberg O. 2000. Fluorescent pigments in corals are photoprotective. Nature 408: 850-853.
-
86. Scatena CD, Hepner MA, Oei YA, Dusich JM, Yu SF, Purchio T, et al. 2004. Imaging of bioluminescent LNCaP-luc-M6 tumors: a new animal model for the study of metastatic human prostate cancer. Prostate 59: 292-303.
-
87. Scott D, Dikici E, Ensor M, Daunert S. 2011. Bioluminescence and its impact on bioanalysis. Annu Rev Anal Chem (Palo Alto Calif) 4: 297-319.
-
88. Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, et al. 2004. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol 21: 841-850.
-
89. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581: 221-224.
-
90. Shen L, Niu J, Wang C, Huang B, Wang W, Zhu N, et al. 2019. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J Virol 93.
-
91. Shimomura O. 1979. Structure of the chromophore of Aequorea green fluorescent protein. FEBS Lett 104: 220-222.
-
92. Shimomura O. 2005. The discovery of aequorin and green fluor- escent protein. J Microsc 217: 1-15.
-
93. Shimomura O. 2012. Bioluminescence: Chemical Principles and Methods. World Scientific., Singapore.
-
94. Shimomura O, Johnson FH. 1978. Peroxidized coelenterazine, the active group in the photoprotein aequorin. Proc Natl Acad Sci U S A 75: 2611-2615.
-
95. Shimomura O, Johnson FH, Saiga Y. 1962. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59: 223-239.
-
96. Shimomura O, Masugi T, Johnson FH, Haneda Y. 1978. Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry 17: 994-998.
-
97. Sparks JS, Schelly RC, Smith WL, Davis MP, Tchernov D, Pieribone VA, Gruber DF. 2014. The covert world of fish biofluorescence: a phylogenetically widespread and phenotypically variable phenomenon. PLoS One 9: e83259.
-
98. Stanger-Hall KF, Lloyd JE, Hillis DM. 2007. Phylogeny of North American fireflies (Coleoptera: Lampyridae): implications for the evolution of light signals. Mol Phylogenet Evol 45: 33-49.
-
99. Stepanyuk GA, Liu ZJ, Markova SS, Frank LA, Lee J, Vysotski ES, Wang BC. 2008. Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.7 A: why it is not a calcium-regulated photoprotein. Photochem Photobiol Sci 7: 442-447.
-
100. Stepanyuk GA, Liu ZJ, Burakova LP, Lee J, Rose J, Vysotski ES, Wang BC. 2013. Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state. Biochim Biophys Acta 1834: 2139-2146.
-
101. Stults NL, Stocks NF, Rivera H, Gray J, McCann RO, O'Kane D, et al. 1992. Use of recombinant biotinylated aequorin in microtiter and membrane-based assays: purification of recombinant apoaequorin from Escherichia coli. Biochemistry 31: 1433-1442.
-
102. Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, et al. 2016. Five colour variants of bright luminescent protein for real-time multicolour bioimaging. Nat Commun 7: 13718.
-
103. Syed AJ, Anderson JC. 2021. Applications of bioluminescence in biotechnology and beyond. Chem Soc Rev 50: 5668-5705.
-
104. Sylvia JM, Janni JA, Klein JD, Spencer KM. 2000. Surface-enhanced raman detection of 2,4-dinitrotoluene impurity vapor as a marker to locate landmines. Anal Chem 72: 5834-5840.
-
105. Takai A, Nakano M, Saito K, Haruno R, Watanabe TM, Ohyanagi T, et al. 2015. Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc Natl Acad Sci U S A 112: 4352-4356.
-
106. Takakura H, Kojima R, Kamiya M, Kobayashi E, Komatsu T, Ueno T, et al. 2015. New class of bioluminogenic probe based on bioluminescent enzyme-induced electron transfer: BioLeT. J Am Chem Soc 137: 4010-4013.
-
107. Takenaka Y, Yamaguchi A, Tsuruoka N, Torimura M, Gojobori T, Shigeri Y. 2012. Evolution of bioluminescence in marine planktonic copepods. Mol Biol Evol 29: 1669-1681.
-
108. Takenaka Y, Masuda H, Yamaguchi A, Nishikawa S, Shigeri Y, Yoshida Y, Mizuno H. 2008. Two forms of secreted and ther- mostable luciferases from the marine copepod crustacean, Metridia pacifica. Gene 425: 28-35.
-
109. Taminiau A, Draime A, Tys J, Lambert B, Vandeputte J, Nguyen N, et al. 2016. HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-kappaB pathway in a transcription-independent manner. Nucleic Acids Res 44: 7331-7349.
-
110. Thompson EM, Nagata S, Tsuji FI. 1989. Cloning and expression of cDNA for the luciferase from the marine ostracod Vargula hilgendorfii. Proc Natl Acad Sci U S A 86: 6567-6571.
-
111. Titushin MS, Markova SV, Frank LA, Malikova NP, Stepanyuk GA, Lee J, Vysotski ES. 2008. Coelenterazine-binding protein of Renilla muelleri: cDNA cloning, overexpression, and character- ization as a substrate of luciferase. Photochem Photobiol Sci 7: 189-196.
-
112. Tsutsui K, Shimada E, Ogawa T, Tsuruwaka Y. 2016. A novel fluor- escent protein from the deep-sea anemone Cribrinopsis japonica (Anthozoa: Actiniaria). Sci Rep 6: 23493.
-
113. Vecchio D, Dai T, Huang L, Fantetti L, Roncucci G, Hamblin MR. 2013. Antimicrobial photodynamic therapy with RLP068 kills methicillin-resistant Staphylococcus aureus and improves wound healing in a mouse model of infected skin abrasion PDT with RLP068/Cl in infected mouse skin abrasion. J Biophotonics 6: 733-742.
-
114. Verhaegent M, Christopoulos TK. 2002. Recombinant Gaussia lucif- erase. Overexpression, purification, and analytical application of a bioluminescent reporter for DNA hybridization. Anal Chem 74: 4378-4385.
-
115. Vysotski ES, Lee J. 2004. Ca2+-regulated photoproteins: structural insight into the bioluminescence mechanism. Acc Chem Res 37: 405-415.
-
116. Wang B, Barahona M, Buck M. 2013. A modular cell-based bio- sensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens Bioelectron 40: 368-376.
-
117. Wang Y, Wang G, O'Kane DJ, Szalay AA. 1998. The Renilla Luciferase-Modified GFP Fusion Protein is Functional in Transformed Cells. BioHydrogen 493-499.
-
-
119. Wehr MC, Rossner MJ. 2016. Split protein biosensor assays in molecular pharmacological studies. Drug Discov Today 21: 415-429.
-
120. Whelan S, Goldman N. 2001. A general empirical model of pro- tein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular Biology and Evolution 18: 691-699.
-
121. White SR, Christopoulos TK. 1999. Signal amplification system for DNA hybridization assays based on in vitro expression of a DNA label encoding apoaequorin. Nucleic Acids Res 27: e25.
-
122. Widder EA. 1999. Bioluminescence. Archer SN, Djamgoz MBA, Loew ER, Partridge JC, Vallerga S (Eds.), Adaptive Mechanisms in the Ecology of Vision. Springer, Dordrecht, pp 555-581.
-
123. Widder EA. 2002. Bioluminescence and the Pelagic Visual Envir- onment. Mar Freshwat Behav Physiol 35: 1-26.
-
124. Widder EA. 2010. Bioluminescence in the ocean: origins of bio- logical, chemical, and ecological diversity. Science 328: 704-708.
-
125. Wood KV, Lam YA, Seliger HH, McElroy WD. 1989. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science 244: 700-702.
-
126. Xiao L, Yang C, Nelson CO, Holloway BP, Udhayakumar V, Lal AA. 1996. Quantitation of RT-PCR amplified cytokine mRNA by aequorin-based bioluminescence immunoassay. J Immunol Methods 199: 139-147.
-
127. Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, et al. 2014. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A 111: 12516-12521.
-
128. Yue JX, Holland ND, Holland LZ, Deheyn DD. 2016. The evolution of genes encoding for green fluorescent proteins: insights from cephalochordates (amphioxus). Sci Rep 6: 28350.
-
129. Zatta PF. 1996. A new bioluminescent assay for studies of protein G and protein A binding to IgG and IgM. J Biochem Biophys Methods 32: 7-13.
-
130. Zeamari S, Rumping G, Floot B, Lyons S, Stewart FA. 2004. In vivo bioluminescence imaging of locally disseminated colon carcinoma in rats. Br J Cancer 90: 1259-1264.
-
131. Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, et al. 2010. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab 11: 364-378.
-
132. Zhao G, Du L, Ma C, Li Y, Li L, Poon VK, et al. 2013. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J 10: 266.